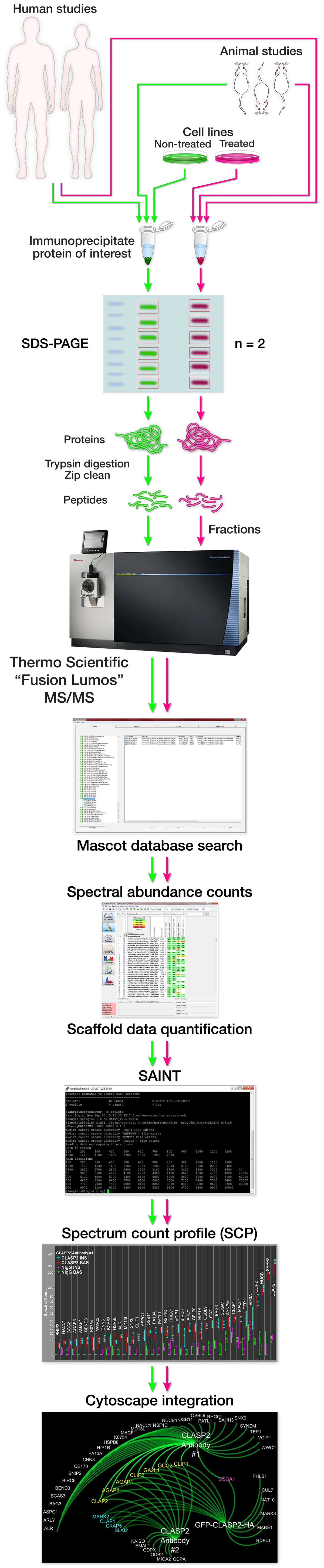
Identifying potential protein interaction partners through characterizing protein interactomes.
If you have a protein of interest and would like to identify potential interaction partners we would like to help. It is our responsibility to aid in the design and execution of the experiment. It begins with a conversation of the specifics and discussion of feasibility, potential complications, and hopeful outcomes. After that, due diligence is performed to ensure efficient "bait" protein purification and yeild. Once appropriate bait protein has been validated a decision is made on the final experimental design and cost. After that, protein lysates from cell culture or animal/human tissue are subjected to bait protein immunoprecipitation and prey protein co-immunoprecipitation as well as negative control immunoprecipitation. The immunoprecipitates are then separated by SDS-PAGE and each gel lane is then fractioned into an appropriated number of gel slices. Each gel slice is then prepped, subjected to enzymatic digestion to digest the protein into peptides, and the resultant peptides are then desalted. Each gel slice's purified peptides are then analyzed as independent samples by mass spectrometry and the mass spec data is then searched against the appropriate database with the software program Mascot. The Mascot data is then put through a second software program Scaffold to allow for easy navigation of the interactome data. The proteins are then scored for enrichment using the bioinformatic software program SAINT and the SAINT-qualified proteins are then assembled into a Spectrum Count Profile figure for easy analysis and presentation. Multiple interactome data can then be integrated using the software program Cytoscape.